Welcome to Next Gen Sequencing
Learn more about Next Generation COVID-19 Sequencing
Professional NGS Covid Testing
With the immediate concern regarding rapidly spreading and increasingly deadly new SARS-CoV-2 coronavirus strains in the US, we must act quickly to ensure the health of US citizens.
Premier Medical Laboratories is expanding our offering of (N)ext (G)eneration (S)equencing capabilities to include Illumina-based COVID-19 Surveillance testing.
NGS Technology Allows us to…
Track the virus transmission routes or hot spots
Detect mutations quickly to stop the spread of new virus strain types (B.1.1.7 & B1.351)
Identify viral mutations that mask, or help targeted strains avoid detection from current molecular diagnostic tests
Screen targets for possible COVID-19 therapeutics
Identify viral mutations that can affect vaccine potency
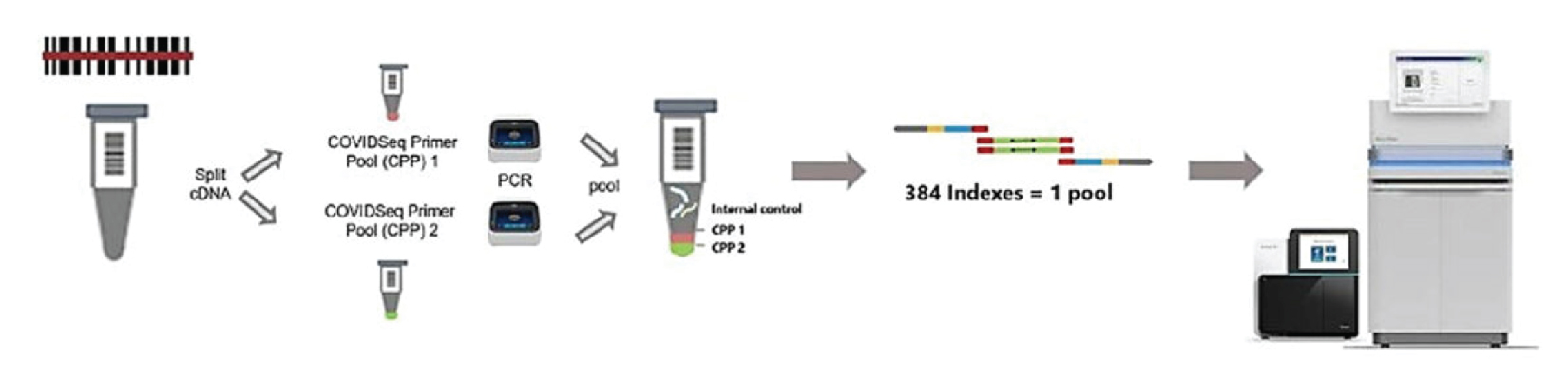
How NGS Technology Works
STEP 1
- RNA extraction
- RNA to CDNA conversion
STEP 2
- Amplify cDNA with COVIDSeq primers (1&2)
- Pool amplicons from primer pool 1 & 2
STEP 3
- Bead linked transposome to fragment and tag amplicons with adapter sequence (includes normalization)
- Amplify tagmented amplicons to add indexes
- Pool libraries by volume
- Washes
STEP 4
- Quantify and dilute library pools
- Load on Sequencer
- Analysis using DRAGEN COVID Lineage
NGS Allows Researchers To:
- Rapidly sequence whole genomes
- Deeply sequence target regions
- Utilize RNA sequencing (RNA-Seq) to discover novel RNA variants and splice sites, or quantify mRNAs for gene expression analysis
- Analyze epigenetic factors such as genome-wide DNA methylation and DNA-protein interactions
- Sequence cancer samples to study rare somatic variants, tumor subclones, and more
- Study the human microbiome
- Identify novel pathogens


Key Features & Benefits

Accurate
Comprehensive
Scalable
Flexible
High-Level Workflow
Manage Workflow
Clarity LIMS Protocols (optional; not part of EUA workflow)
Prepare Library
Illumina COVIDSeq Test
Sequence
NovaSeq 6000 System NextSeq 500/550 Systems NextSeq 550Dx System (in RUO mode)

Analyze
DRAGEN COVIDSeq Test Pipeline (Local) DRAGEN COVIDSeq Test App (BaseSpace Sequencing Hub)
Important Things You Should Know
Questions & Answers
What Is The Sample Collection Method?
This test is authorized for use with nasopharyngeal (NP), oropharyngeal (OP), and mid-turbinate (MT) nasal swabs.
How Many Samples Can Be Run On A NovaSeq 6000 System?
Up to 384 samples can be loaded per lane on a NovaSeq S4 and SP flow cell for a total of 1536 samples per S4 flow cell and 768 per SP flow cell. Two flow cells can be run simultaneously for a total of 3072 or 1536 samples per run respectively.
How Many Samples Can Be Run On NextSeq 500/550/500Dx (In RUO Mode) System?
Up to 384 samples can be loaded per HO flow cell.
How Many Indexes Are Available For This Test?
Up to 384 unique dual indexes (IDT for Illumina PCR Indexes Sets 1-4) are available.
Which Analysis Software Does This Test Use?
Analysis is performed by the DRAGEN COVIDSeq Test Pipeline installed on the local DRAGEN server or by the DRAGEN COVIDSeq Test App on BaseSpace Sequence Hub.
What Does It Mean If The Specimen Tests Positive For SARS-CoV-2?
A positive test result for COVID-19 indicates that RNA from SARS-CoV-2 was detected, and the patient is infected with the virus and presumed to be contagious. Laboratory test results should always be considered in the context of clinical observations and epidemiological data in making a final diagnosis and patient management decisions. Patient management should follow current CDC guidelines.
What Does It Mean If The Specimen Tests Negative For SARS-CoV-2?
A negative test result for this test means that SARS-CoV-2 RNA was not present in the specimen above the limit of detection. However, a negative result does not rule out COVID-19 and should not be used as the sole basis for treatment or patient management decisions. A negative result does not exclude the possibility of COVID-19.
Welcome to NGS
Learn more about Next Generation COVID-19 Sequencing
Professional NGS Covid Testing
With the immediate concern regarding rapidly spreading and increasingly deadly new SARS-CoV-2 coronavirus strains in the US, we must act quickly to ensure the health of US citizens.
Premier Medical Laboratories is expanding our offering of (N)ext (G)eneration (S)equencing capabilities to include Illumina-based COVID-19 Surveillance testing.
NGS Technology Allows us to…
Track the virus transmission routes or hot spots
Detect mutations quickly to stop the spread of new virus strain types (B.1.1.7 & B1.351)
Identify viral mutations that mask, or help targeted strains avoid detection from current molecular diagnostic tests
Screen targets for possible COVID-19 therapeutics
Identify viral mutations that can affect vaccine potency
How NGS Technology Works
STEP 1
- RNA extraction
- RNA to CDNA conversion
STEP 2
- Amplify cDNA with COVIDSeq primers (1&2)
- Pool amplicons from primer pool 1 & 2
STEP 3
- Bead linked transposome to fragment and tag amplicons with adapter sequence (includes normalization)
- Amplify tagmented amplicons to add indexes
- Pool libraries by volume
- Washes
STEP 4
- Quantify and dilute library pools
- Load on Sequencer
- Analysis using DRAGEN COVID Lineage
NGS Allows Researchers To:
- Rapidly sequence whole genomes
- Deeply sequence target regions
- Utilize RNA sequencing (RNA-Seq) to discover novel RNA variants and splice sites, or quantify mRNAs for gene expression analysis
- Analyze epigenetic factors such as genome-wide DNA methylation and DNA-protein interactions
- Sequence cancer samples to study rare somatic variants, tumor subclones, and more
- Study the human microbiome
- Identify novel pathogens

Accurate
Comprehensive
Scalable
Flexible
High-Level Workflow

Manage Workflow
Clarity LIMS Protocols (optional; not part of EUA workflow)

Prepare Library
Illumina COVIDSeq Test

Sequence
NovaSeq 6000 System NextSeq 500/550 Systems NextSeq 550Dx System (in RUO mode)

Analyze
DRAGEN COVIDSeq Test Pipeline (Local) DRAGEN COVIDSeq Test App (BaseSpace Sequencing Hub)
Important Things You Should Know
Questions & Answers
What Is The Sample Collection Method?
This test is authorized for use with nasopharyngeal (NP), oropharyngeal (OP), and mid-turbinate (MT) nasal swabs.
How Many Samples Can Be Run On A NovaSeq 6000 System?
Up to 384 samples can be loaded per lane on a NovaSeq S4 and SP flow cell for a total of 1536 samples per S4 flow cell and 768 per SP flow cell. Two flow cells can be run simultaneously for a total of 3072 or 1536 samples per run respectively.
How Many Samples Can Be Run On NextSeq 500/550/500Dx (In RUO Mode) System?
Up to 384 samples can be loaded per HO flow cell.
How Many Indexes Are Available For This Test?
Up to 384 unique dual indexes (IDT for Illumina PCR Indexes Sets 1-4) are available.
Which Analysis Software Does This Test Use?
Analysis is performed by the DRAGEN COVIDSeq Test Pipeline installed on the local DRAGEN server or by the DRAGEN COVIDSeq Test App on BaseSpace Sequence Hub.
What Does It Mean If The Specimen Tests Positive For SARS-CoV-2?
A positive test result for COVID-19 indicates that RNA from SARS-CoV-2 was detected, and the patient is infected with the virus and presumed to be contagious. Laboratory test results should always be considered in the context of clinical observations and epidemiological data in making a final diagnosis and patient management decisions. Patient management should follow current CDC guidelines.
What Does It Mean If The Specimen Tests Negative For SARS-CoV-2?
A negative test result for this test means that SARS-CoV-2 RNA was not present in the specimen above the limit of detection. However, a negative result does not rule out COVID-19 and should not be used as the sole basis for treatment or patient management decisions. A negative result does not exclude the possibility of COVID-19.
